Molecular Basis Of Inheritance Class 12 Notes For Neet
Nucleic acids (DNA and RNA) are the building blocks of genetic material. DNA is the genetic material in most of the organisms. RNA is the genetic material in some viruses. RNA mostly functions as a messenger.
DNA
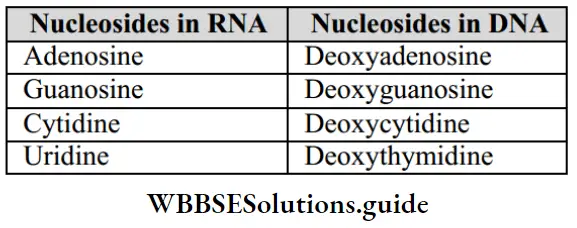
Structure Of Polynucleotide Chain: Polynucleotides are the polymer of nucleotides. DNA and RNA are polynucleotides. A nucleotide has 3 components:
- A nitrogenous base.
- A pentose sugar (ribose in RNA and deoxyribose in DNA).
- A phosphate group.
Nitrogen bases are 2 types:
Read And Learn More: NEET Biology Class 12 Notes
- Purines: These include Adenine (A) and Guanine (G).
- Pyrimidines: It include Cytosine (C), Thymine (T), and Uracil (U). Thymine (5-methyl Uracil) is present only in DNA and Uracil only in RNA.
- A nitrogenous base is linked to the pentose sugar through an N-glycosidic linkage to form nucleoside.
- Nitrogen base + sugar + phosphate group = Nucleotide (deoxyribonucleotide).
- In RNA, every nucleotide residue has an additional -OH group present at the 2′-position in the ribose (2’- OH). 2 nucleotides are linked through 3’-5’ phosphodiester bond to form a dinucleotide. When more nucleotides are linked, it forms polynucleotides.
“molecular basis of inheritance “
Molecular Basis Of Inheritance Class 12 Notes For Neet
Structure Of Thedna
- Friedrich Meischer (1869): Identified DNA and named it as ‘Nuclein’.
- James Watson and Francis Crick (1953) proposed a double helix model of DNA. It was based on X-ray diffraction data produced by Maurice Wilkins and Rosalind Franklin.
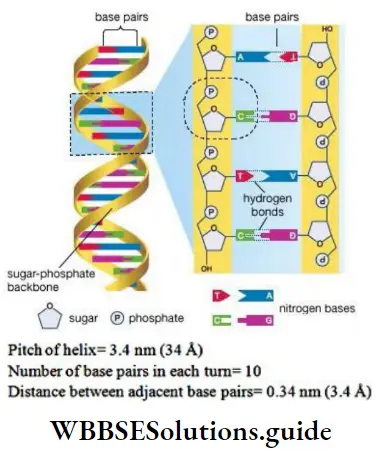
- DNA is made of 2 polynucleotide chains coiled in a right-handed fashion. Its backbone is formed of sugar and phosphates. The bases project inside.
- The 2 chains have anti-parallel polarity, i.e. one chain has the polarity 5’→3’ and the other has 3’→5’.
- The bases in 2 strands are paired through H-bonds forming base pairs (bp).
- A = T (2 hydrogen bonds) C≡G (3 hydrogen bonds)
- Purine comes opposite to a pyrimidine. This generates uniform distance between the 2 strands.
- Erwin Chargaff’s rule: In DNA, the proportion of A is equal to T and the proportion of G is equal to C.
“molecular basis of inheritance notes pdf download “
∴ A [A] + [G] = [T] + [C] or [A] + [G] / [T] + [C] =1
- ∅ 174 (a bacteriophage) has 5386 nucleotides.
- Bacteriophage lambda has 48502 base pairs (bp).
- E. coli has 4.6×106 bp.
- Haploid content of human DNA is 3.3×106 bp.
Length of DNA = number of base pairs X distance between two adjacent base pairs.
The number of base pairs in human = 6.6 x 109

Molecular Basis Of Inheritance Class 12 Notes For Neet
Hence, the length of DNA = 6.6 x 109 x 0.34 x 10-9 = 2.2 m

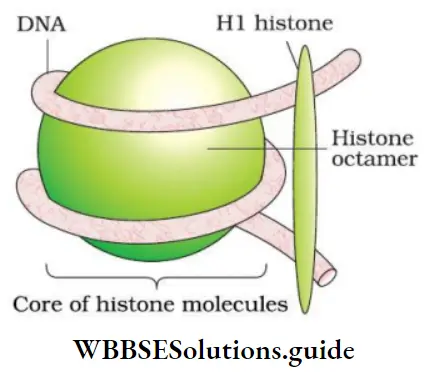
Packaging Of DNA Helix: In prokaryotes (For example, E. coli), the DNA is not scattered throughout the cell. DNA, being negatively charged, is held with some positively charged proteins and forms a ‘nucleoid’. In eukaryotes, there is a set of positively charged, basic proteins called histones. DNA m home
- Histones are rich in positively charged basic amino acid residues lysines and arginines. 8 histones form histone octamer.
- Negatively charged DNA is a Nucleosome wrapped around histone octamer to give nucleosome.
- A typical nucleosome contains 200 bp. Therefore, the total number of nucleosomes in human = \(\frac{6.6 \times 10^9 \mathrm{bp}}{200}=3.3 \times 10^7\)
- Nucleosomes constitute the repeating unit to form chromatin. Chromatin is a thread-like stained body.
- Nucleosomes in chromatin = ‘beads-on-string’.
- Chromatin is packaged → chromatin fibers → coiled and condensed at the metaphase stage → chromosomes.
- Higher level packaging of chromatin requires non-histone chromosomal (NHC) proteins.
NEET Biology Class 12 Molecular Basis of Inheritance Notes
Molecular Basis Of Inheritance Class 12 Notes Neet
“molecular basis of inheritance class 12 pdf notes “
Chromatins include
- Euchromatin: Loosely packed and transcriptionally active chromatin and stains light.
- Heterochromatin: Densely packed and inactive region of chromatin and stains dark.
Search For Genetic Material
1. Griffith’s Transforming Principle experiment Griffith used mice and Streptococcus pneumoniae.
Streptococcus pneumoniae has 2 strains
- Smooth (S) strain (Virulent): Has polysaccharide mucus coat. Cause pneumonia.
- Rough (R) strain (Non-virulent): No mucus coat. Do not cause Pneumonia.
Molecular Basis Of Inheritance Class 12 Notes Neet
Experiment:
- S-strain → Inject into mice → Mice die
- R-strain → Inject into mice → Mice live
- S-strain (Heat killed) → Inject into mice → Mice live
- S-strain (Hk) + R-strain (live) → Inject into mice → Mice die
He concluded that some ‘transforming principle’, transferred the heat-killed S-strain to the R-strain. It enabled the R-strain to smooth the polysaccharide coat and become virulent. due to the transfer of genetic material.
Molecular Basis of Inheritance NEET Notes
2. Biochemical characterization of the transforming principle: Oswald Avery, Colin MacLeod, and Maclyn McCarty worked to determine the biochemical nature of the ‘transforming principle’ in Griffith’s experiment.
- They purified biochemicals (proteins, DNA, RNA, etc.) from heat-killed S cells using suitable enzymes.
- They discovered that
- Digestion of protein and RNA (using Proteases and RNases) did not affect transformation. So, the transforming substance was not a protein or RNA.
- Digestion of DNA with DNase inhibited transformation. It means that DNA caused the transformation of R cells to S cells, i.e. DNA was the transforming principle.
Molecular Basis Of Inheritance Class 12 Notes Neet
3. Hershey-Chase Experiment (Blender Experiment)
- Hershey and Chase grew some bacteriophage viruses on a medium containing radioactive phosphorus (P32) and some others on medium containing radioactive sulphur (S35).
- Viruses grown in P32 got radioactive DNA because only DNA contains phosphorus. Viruses grown in S35 got radioactive protein because protein contains sulfur.
- These preparations were used separately to infect E. coli.
- After infection, the E. coli cells were gently agitated in a blender to remove the virus particles from the bacteria.
- Then the culture was centrifuged to separate lighter virus particles from heavier bacterial cells.
“chapter 6 class 12 biology notes “
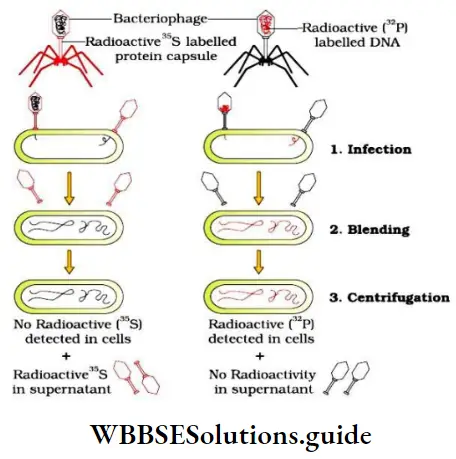
Bacteria infected with viruses have radioactive DNA from radioactive. i.e., DNA had passed from the virus to synthesize Bacteria infected with viruses having radioactive. These must-be proteins were not radioactive. i.e., proteins did not enter the bacteria from the viruses. This proves that DNA is the genetic material.
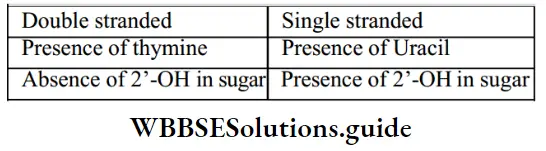
Dna And Rna Difference Class 12
Properties Of Genetic Material DNA Versus RNA
A genetic material may have the following properties:
- Ability to generate its replica (Replication).
- Chemical and structural stability.
- Provide the mutations that are required for evolution.
- Ability to express as ‘Mendelian Characters’.
- RNA is unstable. So, RNA viruses (For example, Q.B bacteriophage, Tobacco Mosaic Virus, etc.) mutate and evolve faster.
- DNA strands are complementary. On heating, they separate. etc) evolved around RNA. In appropriate conditions, they come together. In Griffith’s experiment, some properties of the DNA of the heat-killed bacteria did not destroyed. It indicates the stability of DNA. For the storage of genetic information, DNA is better due to its stability. But for the transmission of genetic information, RNA is better.
- RNA can directly code for protein synthesis, and hence can easily express the characters. DNA is dependent on RNA for protein synthesis.
RNA World
- RNA was the first genetic material.
- It acts as genetic material and catalyst.
- Essential life processes (metabolism, translation, splicing evolved from RNA for stability.
Central Dogma Of Molecular Biology
It is proposed by Francis Crick. It states that the genetic information flows from DNA → RNA → Protein. In some viruses, the flow of information is in the reverse direction (from RNA to DNA). It is called reverse transcription.

Dna And Rna Difference Class 12
DNA Replication
- Replication is the copying of DNA from parental DNA.
- Watson and Crick proposed a Semi-conservative model of replication. It suggests that the parental DNA strands act as templates for the synthesis of new complementary strands. After replication, each DNA molecule would have one parental and one new strand.
- Matthew Messelson and Franklin Stahl (1958) experimentally proved the Semi-conservative model.
NEET Biology Molecular Basis of Inheritance Important Notes
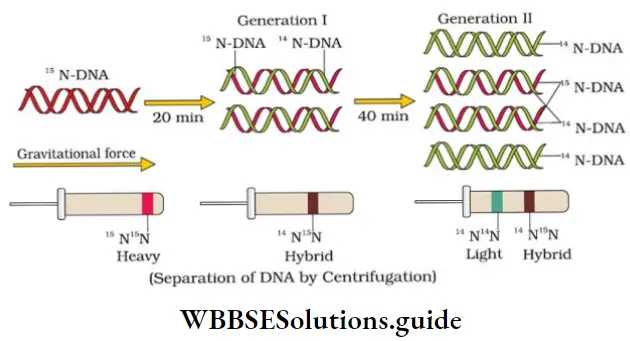
Messelson and Stahl’s Experiment: They cultured E. coli in a medium containing 15NH4Cl (15N: heavy isotope of N). 15N was incorporated into both strands of bacterial DNA and the DNA became heavier.
- Another preparation containing N salts labeled with 14N is also made. 14N was incorporated in both strands of DNA and became lighter. These 2 types of DNA can be separated by centrifugation in a CsCl density gradient.
- They took E. coli cells from 15N medium and transferred to 14N medium. After one generation (i.e. after 20 minutes), they isolated and centrifuged the DNA. Its density was intermediate (hybrid) between 15N DNA and 14N DNA. This shows that the newly formed DNA one strand is old (15N type) and one strand is new (14N type). This confirms semi-conservative replication.
- After 2 generations (i.e. after 40 minutes), there was equal amounts of hybrid DNA and light DNA.
- Taylor and colleagues (1958) performed similar experiments on Vicia faba (faba beans) using radioactive thymidine to detect distribution of newly synthesized DNA in the chromosomes. It proved that the DNA in chromosomes also replicate semi-conservatively.
“molecular basis of inheritance pyq neet “
Dna And Rna Difference Class 12
The Machinery and Enzymes for Replication
- DNA replication starts at a point called origin (ori). A unit of replication with one origin is called a replicon.
- During replication, the 2 strands unwind and separate by breaking H-bonds in presence of an enzyme, Helicase. Unwinding of the DNA molecule at a point forms a ‘Y’-shaped
- The separated strands act as templates for the synthesis of new strands. DNA replies in the 5’→3’ direction.
- Deoxyribonucleoside triphosphates (dATP, dGTP, dCTP, and dTTP) act as substrate and provide energy for polymerization.
- Firstly, a small RNA primer is synthesized in presence of an enzyme, primase.
- In the presence of an enzyme, DNA-dependent DNA polymerase, many nucleotides join with one another to primer strand and form a polynucleotide chain (new strand).
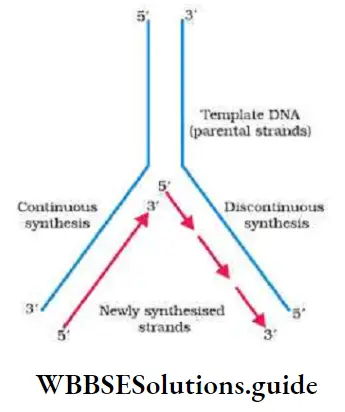
- The DNA polymerase forms one new strand (leading strand) in a continuous stretch in the 5’^3’ direction (Continuous synthesis).
- The other new strand is formed in small stretches (Okazaki fragments) in 5’→3’ direction (Discontinuous synthesis).
- The Okazaki fragments are then joined together to form a new strand by an enzyme, DNA ligase. This new strand is called lagging strand.
- If a wrong base is introduced in the new strand, DNA polymerase can do proof reading. E. coli completes replication within 18 minutes. i.e. 2000 bp per second.
- In eukaryotes, the replication of DNA takes place at the S- phase of the cell cycle. Failure in cell division after DNA replication results in polyploidy.
Molecular Basis of Inheritance Class 12 NEET Key Concepts and Summary
Transcription
It is the process of copying genetic information from one strand of the DNA into RNA. Here, adenine pairs with uracil instead of thymine. During transcription, both strands are not copied because The code for proteins is different in both strands. This complicates the translation.
If 2 RNA molecules are produced simultaneously, this would be complimentary to each other. It forms a double-stranded RNA and prevents translation.
Class 12 Biology Molecular Basis Of Inheritance Notes
Transcription Unit
- It is the segment of DNA between the sites of initiation and termination of transcription. It consists of 3 regions:
- A promoter (Transcription start site): Binding site for RNA polymerase.
- Structural gene: The region between promoter and terminator where transcription takes place.
- A terminator: The site where transcription stops.
- The DNA-dependent RNA polymerase catalyzes the polymerization only in 5’ → 3’direction.
- 3’→5’ acts as template strand. 5’→3’ acts as a coding strand. ATGCATGCATGCATGCATGCATGC-5’te mplate strand. TACGTACGTACGTACGTA CGTACG-3’ coding strand.
“bank of biology class 12 molecular basis of inheritance “
Transcription unit and gene: A gene is a functional unit of inheritance. It is the DNA sequence coding for an RNA (mRNA, rRNA, or tRNA). Cistron is a segment of DNA coding for a polypeptide during protein synthesis. It is the largest element of a gene. Structural gene in a transcription unit is 2 types:
- Monocistronic structural genes (split genes): It is seen in eukaryotes. Here, coding sequences (exons or expressed sequences) are interrupted by introns (intervening sequences). Exon sequences appear in processed mRNA. Intron sequences do not appear in processed mRNA.
- Polycistronic structural genes: It is seen in prokaryotes. Here, there are no split genes.
Steps of transcription in prokaryotes: In bacteria (Prokaryotes), synthesis of all types of RNA are catalysed by a single RNA polymerase. It has 3 steps:
- Prokaryotes Initiation: Here, the enzyme RNA polymerase binds at the promoter site of DNA. This causes the local unwinding of the DNA double helix. An initiation factor(factor) present in RNA polymerase initiates RNA synthesis.
- Prokaryotes Elongation: RNA chain is synthesized in 5’ → 3’ direction. In this process, activated ribonucleoside triphosphates (ATP, GTP, UTP, and CTP) are added. This is complementary to the base sequence in the DNA template.
- Prokaryotes Termination: A termination factor (p factor) binds to the RNA polymerase and terminates the transcription.
- mRNA requires no processing to become active.
- Transcription and translation take place in the same compartment (no separation of cytosol and nucleus).
Class 12 Biology Molecular Basis Of Inheritance Notes
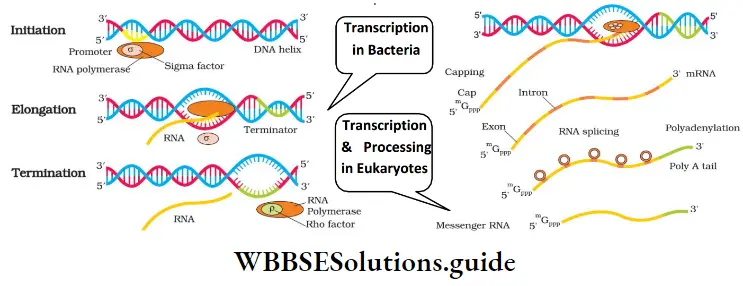
In eukaryotes, there are 2 additional complexities:
1. There are 3 RNA polymerases:
- RNA polymerase 1: Transcribes rRNAs (28S, 18S and 5.8S).
- RNA polymerase 2: Transcribes the heterogeneous nuclear RNA (hnRNA). It is the precursor of mRNA.
- RNA polymerase 3: ‘Transcribes tRNA, 5S rRNA, and snRNAs (small nuclear RNAs).
2. The primary transcripts (hnRNA) contain exons and introns and are non-functional. Hence introns must be removed. For this, it undergoes the following processes:
Class 12 Biology Molecular Basis Of Inheritance Notes
- Splicing: From hnRNA introns are removed (by the spliceosome) and exons are spliced (joined) together.
- Capping: Here, a nucleotide methyl guanosine triphosphate (cap) is added to the 5’ end of hnRNA.
- Tailing (Polyadenylation): Here, adenylate residues (200-300) are added at 3’-end. It is the fully processed hnRNA, now called mRNA.
Genetic Code
It is the sequence of nucleotides (nitrogen bases) in mRNA that contains information for protein synthesis (translation). The sequence of 3 bases determining a single amino acid is called codon.
George Gamow suggested that for coding 20 amino acids, the code should be made up of 3 nucleotides. Thus, there are 64 codons (43= 4 x 4 x 4).
- Har Gobind Khorana developed the chemical method in synthesizing RNA molecules with defined combinations of bases (homopolymers and copolymers).
- Marshall Nirenberg developed a cell-free system for protein synthesis.
- Severo Ochoa (polynucleotide phosphorylase) enzyme is used to polymerize RNA with defined sequences in a template-independent manner.
NEET Biology Class 12 Chapter Molecular Basis of Inheritance Detailed Notes
20 types of amino acids involved in translation
- Alanine (Ala)
- Arginine (Arg)
- Asparagine (Asn)
- Aspartic acid (Asp)
- Cystein (Cys)
- Glutamine (Gln)
- Glutamic acid (Glu)
- Glycine (Gly)
- Histidine (His)
- Isoleucine (Ile)
- Leucine(Leu)
- Lysine (Lys)
- Methionine(Met)
- Phenylalanine(Phe)
- Proline (Pro)
- Serine (Ser)
- Threonine (Thr)
- Tryptophan (Trp)
- Tyrosine (Tyr)
- Valine (Val)
The codons for the various amino acids
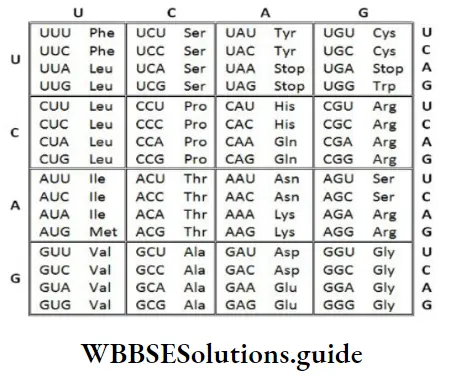
Salient features of genetic code
Class 12 Biology Molecular Basis Of Inheritance Notes
- Triplet code (three-letter code).
- 61 codons code for amino acids. 3 codons (UAA, UAG, and UGA) do not code for any amino acids. They act as stop codons (Termination codons or non-sense codons). Genetic code is universal. Example, From bacteria to human UUU codes for Phenylalanine. Some exceptions are found in mitochondrial codons, and in some protozoans.
- No punctuations b/w adjacent codons (comma less code). The codon is read in mRNA in a contiguous fashion.
- Genetic code is non-overlapping.
- A single amino acid is represented by many codons (except AUG for methionine and UGG for tryptophan). Such codons are called degenerate codons.
- Genetic code is unambiguous and specific. i.e. one codon specifies only one amino acid.
- AUG has dual functions. It codes for Methionine and acts as an initiator codon. In eukaryotes, methionine is the first amino acid, and formyl methionine in prokaryotes.
Biology Class 12 Notes For Neet
Mutations and Genetic Code: The relationship between genes and DNA are best understood by mutation studies. Deletions and rearrangements in DNA may cause the loss or gain of a gene and so a function.
- Insertion or deletion of one or two bases changes the reading frame from the point of insertion or deletion. Insertion/ deletion of three or its multiple bases insert or delete one or multiple codons.
- Hence one or multiple amino acids are inserted /deleted. The reading frame remains unaltered from that point onwards. Such mutations are known as frame-shift insertion or deletion mutations. It proves that codon is a triplet and is read contiguously.
Types Of RNA
- mRNA (messenger RNA): Provide template for translation (protein synthesis).
- rRNA (ribosomal RNA): Structural and catalytic role during translation. For example, 23S rRNA in bacteria acts as ribozyme.
- tRNA (transfer RNA or sRNA or soluble RNA): Brings amino acids for protein synthesis and reads the genetic code.
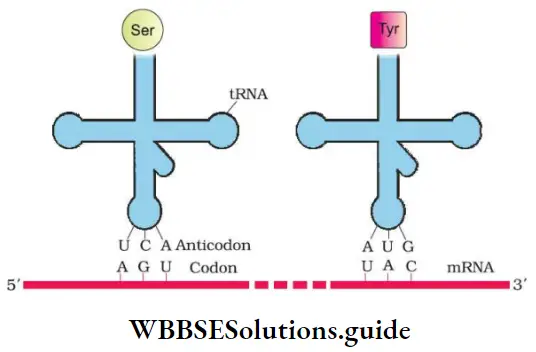
NEET Study Material for Molecular Basis of Inheritance Chapter
Francis Crick postulated presence of an adapter molecule that can read the code and to link with amino acids. tRNA is called adaptor molecule because it has
Biology Class 12 Notes For Neet
- An Anticodon (NODOC) loop that has bases complementary to the codon.
- An amino acid acceptor end to which amino acid binds.
- Ribosome binding loop.
- Enzyme binding loop.
- For initiation, there is another tRNA called initiator tRNA. There are no tRNAs for stop codons.
- The secondary (2-D) structure of tRNA looks like a cloverleaf. The 3-D structure looks like an inverted ‘L’.
Translation (Protein Synthesis)
It is the process of polymerization of amino acids to form a polypeptide based on the sequence of codons in mRNA.
“molecular basis of inheritance class 12 notes bank of biology “
It takes place in ribosomes. It includes 4 steps:
- Charging of tRNA
- Initiation
- Elongation
- Termination
1. Charging (aminoacylation) of tRNA: Formation of peptide bond needs energy obtained from ATP. For this, amino acids are activated (amino acid + ATP) and linked to their cognate tRNA in the presence of aminoacyl tRNA synthetase. Thus, the tRNAbecomes charged.
Biology Class 12 Notes For Neet
2. Initiation
- It begins at the 5 ’-end of mRNA in the presence of an initiation factor. The mRNA binds to the small subunit of ribosome. Now the large subunit binds to the small subunit to complete the initiation complex.
- Large subunit has 2 binding sites for tRNA- aminoacyl tRNA binding site (A site) and peptidyl site (P site). The initiation codon for methionine is AUG. So methionyl tRNA complex would have UAC at the Anticodon site.
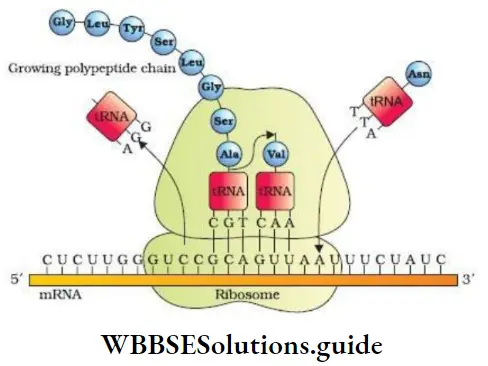
Molecular Basis of Inheritance Class 12 NCERT Notes for NEET
3. Elongation
- At the P site, the first codon of mRNA binds with anticodon of methionyl tRNA complex.
- Another aminoacyl tRNA complex with an appropriate amino acid enters the ribosome and attaches to A site. Its anticodon binds to the second codon on the mRNA and a peptide bond is formed between the first and second amino acids in the presence of an enzyme, peptidyltransferase.
- First amino acid and its tRNA are broken. This tRNA is removed from P site and the second tRNA at the A site is pulled to P site along with mRNA. This is called translocation.
- Then 3rd codon comes into A site and a suitable tRNA with 3rd amino acid binds at the A site. This process is repeated. During translation, ribosome moves from codon to codon.
Biology Class 12 Notes For Neet
4. Termination
- When a release factor binds to the stop codon, the translation terminates. The polypeptide and tRNA are released from the ribosomes.
- The ribosome dissociates into large and small suClass 12 Biology Notes For Neetbunits at the end of protein synthesis. A group of ribosomes associated with a single mRNA for translation is called a polyribosome (polysomes).
- An mRNA has additional sequences that are not translated (untranslated regions or UTR). UTRs are present at both 5’-end (before the start codon) and 3’-end (after the stop codon). They are required for efficient translation process.
Regulation Of Gene Expression
In eukaryotes gene expression occurs by following levels.
- Transcriptional level (formation of primary transcript).
- Processing level (splicing etc.).
- Transport of mRNA from nucleus to the cytoplasm.
- Translational level (formation of apolypeptide).
Class 12 Biology Notes For Neet
The metabolic, physiological, and environmental conditions regulate the expression of genes. Example,
- In E. coli, the beta-galactosidase enzyme hydrolyses lactose into galactose and glucose. In the absence of lactose, the synthesis of beta-galactosidase stops.
- The development and differentiation of embryos into adult are a result of the expression of several set of genes. a substrate is added to the growth medium of bacteria, a set of switched off on metabolize it is called induction.
- When a metabolite (product) is added, the genes to produce it are turned off. This is called repression.
Operon Concept
- “Each metabolic reaction is controlled by a set of genes”
- All the genes regulating a metabolic reaction constitute an protein. For example, lac operon, trp operon, ara operon, his becomes free operon, etc.
Class 12 Biology Notes For Neet
Lac Operon in E. coli
- The operon controlling lactose metabolism
- It is proposed by Francois Jacob and Jacque Monod. It consists of
- A regulatory or inhibitor
- gene: Codes for the repressor.
- 3 structural genes:
- z gene: Codes for β galactosidase (hydrolyze lactose to galactose and glusoce.
- y gene: Codes for permease (increase the permeability of the cell to lactose).
- a gene: Codes for a transacetylase.
The genes present in the operon function together in the same or related metabolic pathway. There is an operator region for each operon. If there is no lactose (inducer), lac operon remains If it is switched off. The regulator gene synthesizes mRNA to news is produce the repressor protein. This protein binds to the operator genes and blocks RNA polymerase movement. So the structural genes are not expressed.
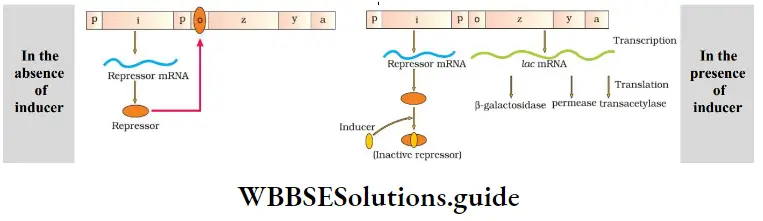
DNA Replication, Transcription, and Translation NEET Notes
If lactose is provided in the growth medium, the lactose is transported into the E. coli cells by the action of permease Lactose (inducer) binds with repressor protein. So repressor protein cannot bind to the operator gene. The operator gene is free and induces the RNA polymerase to bind with the operon, val promoter gene. Then transcription starts. Regulation of lac operon by repressor is called negative regulation.
Human Genome Project
The entire DNA in the haploid set of chromosomes of an organism is called a Genome. In Human genome, DNA is packed in 23chromosomes
- Human genome contains about 3x109bp.
- Human Genome Project (1990-2003) was the first mega project in identifying the sequence of nucleotides and mapping of all the genes in human genome.
- HGP was coordinated by U.S. Department of Energy and the National Institute of Health.
Class 12 Biology Notes For Neet
Goals of HGP
- Identify all the estimated genes in human DNA.
- Determine the sequences of the 3 billion chemical base pairs that make up humanDNA.
- Store this information in databases.
- Improve tools for data analysis.
- Transfer related technologies to other sectors. Address the ethical, legal, and social issues (ELSI) that may arise from the project.
Methodologies of HGP: 2 major approaches.
- Expressed Sequence Tags (ESTs): Focused on identifying all the genes that are expressed as RNA.
- Sequence annotation: Sequencing whole set of genome containing all the coding and non-coding sequence and later assigning different regions in the sequence with functions.
HGP Procedure of sequencing: Isolate total DNA from a cell → Convert into random fragments → Clone in suitable host (bacteria and yeast) using vectors (for example, BAC and YAC) for amplification → Fragments are sequenced using Automated DNA sequencers (using Frederick Sanger method) → Sequences are arranged based on overlapping regions → Alignment of sequences using computer programs.
- BAC= Bacterial Artificial Chromosomes
- YAC= Yeast Artificial Chromosomes
“molecular basis of inheritance class 12 notes bank of biology “
DNA is converted to random fragments as there are technical limitations in sequencing very long pieces of DNA. HGP was closely associated with Bioinformatics. Bioinformatics: Application of computer science and information technology to the field of biology and medicine.
DNA sequencing also have been done in bacteria, yeast, Caenorhabditis elegans (a free living non-pathogenic nematode), Drosophila, plants (rice and Arabidopsis), etc.
Class 12 Biology Notes For Neet
Salient Features of the Human Genome
- The human genome contains 3164.7 million nucleotide bases.
- Total number of genes= about 30,000.
- Average gene consists of 3000 bases, but sizes vary. Largest known human gene (dystrophin on X- X-chromosome) contains 2.4 million bases.
- 99.9% nucleotide bases are same in all people. Only a 0.1% (3×106 bp) difference makes every individual unique.
- Functions of over 50% of discovered genes are unknown.
- Chromosome I has most genes (2968) and Y has the fewest (231).
- Less than 2% of the genome codes for proteins.
- A very large portion of the human genome is made of Repeated (repetitive) sequences. These are stretches of DNA sequences that are repeated many times. They have no direct coding functions. They shed light on chromosome structure, dynamics and evolution.
- About 1.4 million locations have single-base DNA differences. They are called SNPs (Single nucleotide polymorphism or ‘snips’). This helps to find chromosomal locations for disease-associated sequences and tracing human history.
DNA Fingerprinting (DNA Profiling)
It is the technique to identify the similarities and differences of the DNA fragements of 2 individulas. Developed by Alec Jeffreys (1985).
Basis of DNA fingerprinting
Class 12 Biology Notes For Neet
- DNA carries some non-coding repetitive sequences called variable number tandem repeats (VNTR).
- Number of repeats is specific from person to person.
- The size of VNTR varies from 0.1 to 20 kb.
- Repetitive DNA are separated from bulk genomic DNA as different peaks during density gradient centrifugation.
- The bulk DNA forms a major peak and the small peaks
called satellite DNA. - Satellite DNA is classified as micro-satellites, mini¬satellites, etc. based on base composition (A: T rich or G: C rich), length of the segment, and number of repetitive units.
- VNTR belongs to mini-satellite DNA.
- Any difference in the nucleotide sequence (inheritable mutation) observed in a population is called DNA polymorphism (variation at genetic level).
- Polymorphism is higher in non-coding DNA sequence because mutations in these sequences may not have any immediate effect in an individual’s reproductive ability. These mutations accumulate generation after generation and cause polymorphism.
- Polymorphisms have great role in evolution and speciation differences of the
Steps of DNA fingerprinting (Southern Blotting Technique)
- Isolation of DNA.
- Digestion of DNA by restriction endonucleases.
- Separation of DNA fragments by gel electrophoresis.
- Transferring (blotting) DNA fragments to synthetic membranes such as nitrocellulose or nylon.
- Hybridization using radioactively labeled VNTR probe.
- Detection of hybridized DNA by autoradiography.
“molecular basis of inheritance class 12 notes bank of biology “
The image (in the form of dark and light bands) obtained is are called DNA fingerprint. It differs in everyone except in monozygotic (identical) twins.
The sensitivity of the technique can be increased by use of polymerase chain reaction (PCR). Therefore, DNA from a single cell is enough for DNA fingerprinting.
Application of DNA fingerprinting
- Forensic tool to solve paternity, rape, murder etc.
- For the diagnosis of genetic diseases.
- To determine phylogenetic status of animals.
- To determine population and genetic diversities.
